I take an almost unhealthy pleasure in pushing my computer to its limits. This has become easier with Revolution R and its free license for academic use. One of its best features is debugger that allows you to step through R code interactively like you can with python on PyDev. The other useful thing it packages is a simple way to run embarrassingly parallel jobs on a multicore box with the doSMP package.
library(doSMP)
# This declares how many processors to use.
# Since I still wanted to use my laptop, during the simulation I chose cores-1.
workers <- startWorkers(7)
registerDoSMP(workers)
# Make Revolution R not try to go multi-core since we're already explicitly running in parallel
# Tip from: http://blog.revolutionanalytics.com/2010/06/performance-benefits-of-multithreaded-r.html
setMKLthreads(1)
chunkSize <- ceiling(runs / getDoParWorkers())
smpopts <- list(chunkSize=chunkSize)
#This just let's me see how long the simulation ran
beginTime <- Sys.time()
#This is the crucial piece. It parallelizes a for loop among the workers and aggregates their results
#with cbind. Since my function returns c(result1, result2, result3), r becomes a matrix with 3 rows and
# "runs" columns.
r <- foreach(icount(runs), .combine=cbind, .options.smp=smpopts) %dopar% {
# repeatExperiment is just a wrapper function that returns a c(result1, result2, result3)
tmp <- repeatExperiment(N,ratingsPerQuestion, minRatings, trials, cutoff, studentScores)
}
runTime <- Sys.time() - beginTime
#So now I can do something like this:
boxplot(r[1,], r[2,], r[3,],
main=paste("Distribution of Percent of rmse below ", cutoff,
"\n Runs=", runs, " Trials=",trials, " Time=",round(runTime,2)," mins\n",
"scale: ",scaleLow,"-",scaleHigh,
sep=""),
names=c("Ave3","Ave5","Ave7"))
If you are intersested in finding out more of about this, their docs are pretty good.
The only drawback is that Revolution R is a bit rough around the edges and crashes much more than it should. Worse, for me at least the support forum doesn’t show any posts when I’m logged in and I can’t post anything. Although I’ve filled out (what I think is) the appropriate web-form no one has gotten back to me about fixing my account. I’m going to try twitter in a bit. Your mileage may vary.Update: 6/9/2010 22:03 EST
Revolution Analytics responded to my support request after I mentioned it on twitter. Apparently, they had done something to the forums which corrupted my account. Creating a new account fixed the problem, so now I can report the bugs that I
find and get some help.
Update: 6/11/2010 16:03 EST
It turns out that you get a small speed improvement by setting setMKLthreads(1). Apparently, the libraries Revolution R links against attempt to use multiple cores by default. If you are explicitly parrallel programing, this means that your code is competing with itself for resources. Thanks for the tip!

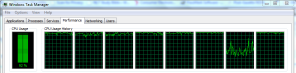
Nathan:
I saw on this blog post (http://blog.revolutionanalytics.com/2010/06/pegging-7-cores-with-dosmp.html) that you have 8 cores on a laptop? Can you point out which laptop as I am in the market this year for a new laptop with as many cores as possible for use with R. I have a feeling what David Smith meant was an 8 core computer. Thanks!
Thanks for the example!
I would be curious how, performance-wise, your simulation would run using some other parallel framework in R. For example the ‘multicore’ package. You could do it with something like:
library(multicore)
# matrix of simulation conditions
conditions <- matrix(rnorm(900), 100, 3)
# single simulation run
doit <- function(i)
{
# vector of inputs
v <- conditions[i,]
# some dummy calculations
c( result1=(v[1] + v[2]) * v[3],
result2=(v[2] + v[3]) * v[1],
result3=(v[3] – v[2]) * v[1] )
}
# this gives a list with a component for every run,
# change mc.cores to 8 in your setup
l <- mclapply(seq(1, nrow(conditions)), doit, mc.cores=3)
# cbind it
r <- do.call("cbind", l)
I’m praying to R gods every evening to implement multicore to Windows as well.
@vinh I have an Asus N61JQ-X1. I chose it by getting the CPU performance ranking from http://www.cpubenchmark.net/cpu_list.php and then comparing on price to find the cheapest processing power at the 1k price point. It’s great, but the battery only lasts for two hours.
@michal
I’ll compare when I return from the IES conference and report back. Thanks for the sample to check with!
@nathanvan Thanks for your response. Looks to me like the i7 processor is still a quad core. It might show up as 8 logical cores due to the multi-threading? Can you confirm?
Sorry to bother you with such requests, but can you try your code with 8 cores and 4 cores, and see if there really is an advantage? If there is, try it with 10 cores specified. I’m convinced that specifying 8 cores can’t beat 4 since the computer is a quad-core. I hope to be proven otherwise (I’m interested because, as said before, I’m in the market for a laptop I can do parallel computing with R).
Also, although its a quad-core, the clock speed is at 1.8ghz. Can you comment on the system performance? Regular usage and when computing with R? I’d like to know whether 1.8ghz suffices or whether I should really go for something with a higher clock speed like 2.4ghz (quad). Thanks!
@vinh Yeah, I’m being sloppy. It’s quad core w/ two threads per core: http://ark.intel.com/Product.aspx?id=43122.
Changing from 8 to 4 would cut the performance roughly in half depending on the communication overhead; in this example it should be pretty close to half. I’ll run it when I do michal’s stuff and make a separate post.
It’s fast. I like it. Windows 7 works well; Ubuntu 10.04 is much faster. Depends on what you can afford and what you want I guess. 🙂 Seriously though, check out the passmark site. It will help you judge how much processing power you are buying.
@nathanvan Thanks again for the response. Will look more into passmark. I do plan to load Ubuntu on it.
Can you post a comment to this post when you post a new post on the 4 core, 8 core, and 10 core test? I’d like to know the results, thanks!
I foundTal’s contribution on his r-statistics blog very helpful:
http://www.r-statistics.com/2010/04/parallel-multicore-processing-with-r-on-windows/
@michal Since I haven’t setup revolution R on linux yet, I’m not able to test out the performance of other multi-core packages that require linux. Maybe once I get a little farther on my actual work I’ll be able to come back to it.
For those of you that haven’t seen this yet, Ryan Rosario (@DataJunkie), gave a talk that is much better than anything that I’ve been able to do: http://www.bytemining.com/2010/07/taking-r-to-the-limit-part-i-parallelization-in-r/
Sure, I was just curious…
As a matter of fact, you don’t need RevoR to run the example I gave you. Base R plus ‘multicore’ package (plus any extras you need for your simulation) is enough.
Unless, of course, you meant that you simply don’t have linux machine available…
@michal Right. I am worried that any difference between doSMP running RevoR on windows and multicore running vanilla R on linux might be easily explained by the different scheduling approaches of the two operating systems. (For example the X264 kernel thing: http://x264dev.multimedia.cx/?p=185) Hence the desire to compare both on linux. And then revo vs vanilla. And then compare with windows performance.
I am interested in finding the answer. Once I get back to the simulation portion of my project I might squeeze some time into to do the experiments. 🙂